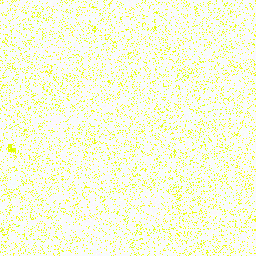| 0. Reference |
0° |
255,128,128 |
34,313,968 |
Composite of all amino acids |
 |
| 1. Histidine |
329° |
255,128,193 |
1,147,212 |
Group IV: Basic amino acids |
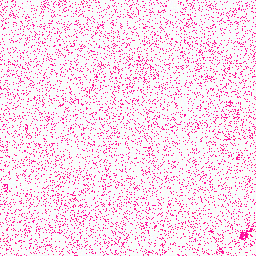 |
| 2. Glutamic acid |
16° |
255,162,128 |
1,268,506 |
Group III: Acidic amino acids |
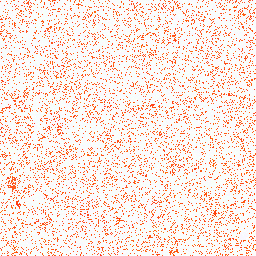 |
| 3. Aspartic acid |
31° |
255,193,128 |
781,982 |
Group III: Acidic amino acids |
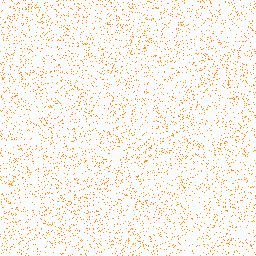 |
| 4. Lysine |
313° |
255,128,227 |
2,010,980 |
Group IV: Basic amino acids |
 |
| 5. Cysteine |
63° |
249,255,128 |
1,194,135 |
Group II: Polar, uncharged amino acids |
 |
| 6. Glycine |
78° |
217,255,128 |
1,779,960 |
Group I: Nonpolar amino acids |
 |
| 7. Alanine |
94° |
183,255,128 |
1,457,198 |
Group I: Nonpolar amino acids |
 |
| 8. Valine |
125° |
128,255,138 |
1,731,907 |
Group I: Nonpolar amino acids |
 |
| 9. Leucine |
141° |
128,255,172 |
3,765,793 |
Group I: Nonpolar amino acids |
 |
| 10. Isoleucine |
157° |
128,255,206 |
1,999,041 |
Group I: Nonpolar amino acids |
 |
| 11. Phenylalanine |
172° |
128,255,238 |
1,990,701 |
Group I: Nonpolar amino acids |
 |
| 12. Tryptophan |
188° |
128,238,255 |
634,648 |
Group I: Nonpolar amino acids |
 |
| 13. Serine |
203° |
128,206,255 |
3,077,813 |
Group II: Polar, uncharged amino acids |
 |
| 14. Threonine |
219° |
128,172,255 |
1,732,854 |
Group II: Polar, uncharged amino acids |
 |
| 15. Glutamine |
250° |
149,128,255 |
1,350,696 |
Group II: Polar, uncharged amino acids |
 |
| 16. Asparagine |
266° |
183,128,255 |
1,361,436 |
Group II: Polar, uncharged amino acids |
 |
| 17. Tyrosine |
282° |
217,128,255 |
1,085,469 |
Group II: Polar, uncharged amino acids |
 |
| 18. Arginine |
297° |
249,128,255 |
1,702,842 |
Group IV: Basic amino acids |
 |
| 19. Proline |
344° |
255,128,162 |
1,773,747 |
Group I: Nonpolar amino acids |
 |
| 20. Methionine |
110° |
149,255,128 |
639,778 |
START Codon |
 |
| 21. Ochre |
0° |
255,128,128 |
709,183 |
STOP Codon |
 |
| 22. Amber |
47° |
255,227,128 |
442,672 |
STOP Codon |
 |
| 23. Opal |
240° |
128,128,255 |
675,415 |
STOP Codon |
 |